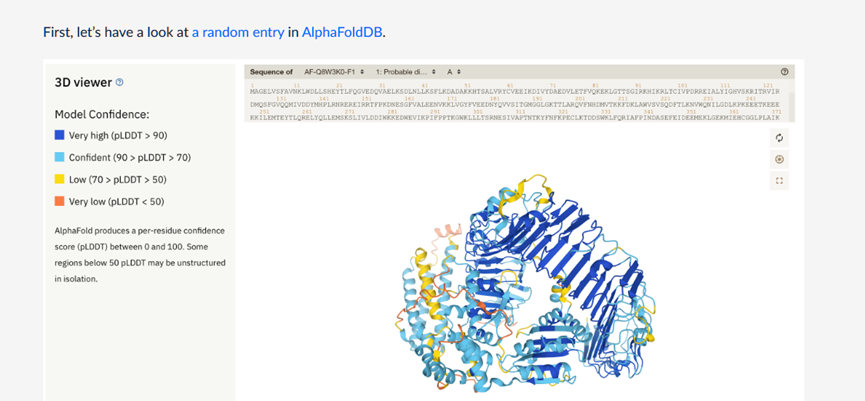
color by b_factor
AF2样图1
代码1-分块
- #将b_factor对应到每个氨基酸残基上
- from biopandas.pdb import PandasPdb
- file = '/home/databank/lywu/vp37/af2-vp37/ranked_0.pdb'
- data = PandasPdb().read_pdb(file)
- #data.df.keys()
- #data.df['ATOM'].columns
- atom_df = data.df['ATOM']
- #根据氨基酸编号进行去重,并重设Index
- residue_list = atom_df.drop_duplicates(subset=['residue_number']).reset_index(drop=True)
- #输出去重后每个氨基酸对应的b_factor
- b_factor_list = residue_list['b_factor']
- '''
- 根据筛选条件,输出对应的氨基酸编号,注意+1
- '''
- #very low
- filter1 = np.where([(b_factor_list < 50)])[1]
- #low
- filter2 = np.where([(b_factor_list >= 50)&(b_factor_list < 70)])[1]
- #Confident
- filter3 = np.where([(b_factor_list >= 70)&(b_factor_list < 90)])[1]
- #very high
- filter4 = np.where([(b_factor_list >= 90)])[1]
- #根据b_factor分类对残基重新着色
- from pymol import cmd
- cmd.delete("all")
- cmd.load(file)
- color_list = ['orange','yellow','palecyan', 'skyblue']
- filetr_list = [filter1, filter2, filter3, filter4]
- for i in range(4):
- colorid = color_list[i]
- f_index = filetr_list[i]
- for index in f_index:
- resid = index + 1
- cmd.color(colorid,"resi " + str(resid))
- cmd.save("test2.pse")
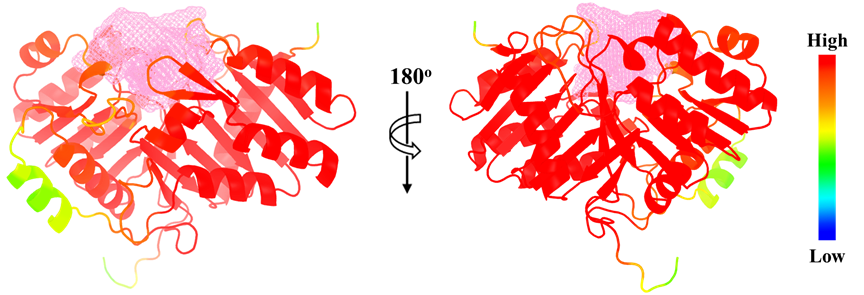
样图2
代码2-连续
b_factor的归一化处理
- from biopandas.pdb import PandasPdb
- file = '/home/databank/lywu/vp37/af2-vp37/ranked_0.pdb'
- data = PandasPdb().read_pdb(file)
- data.df.keys()
- data.df['ATOM'].columns
- df = data.df['ATOM'].b_factor
- df_normalized = df.apply(lambda x:(x - 0)/(100 - 0))
- #df_normalized此时对应的是单个原子,需要另外处理时期对应每个残基
绘制colorbar
使用默认的colormap
- fig, ax = plt.subplots(figsize=(1, 6))
- fig.subplots_adjust(bottom=0.5)
- cmap = mpl.cm.Pastel1
- norm = mpl.colors.Normalize(vmin=40, vmax=100)
- fig.colorbar(mpl.cm.ScalarMappable(norm=norm, cmap=cmap),
- cax=ax, orientation='vertical', label='Some Units')
自定义colormap, 即colorlist
- from matplotlib import colors
- fig, ax = plt.subplots(figsize=(1, 6))
- fig.subplots_adjust(bottom=0.5)
- colorslist = ['#104E8B','white']
- cmap = colors.LinearSegmentedColormap.from_list('mylist',colorslist,N=10)
- norm = mpl.colors.Normalize(vmin=40, vmax=100)
- fig.colorbar(mpl.cm.ScalarMappable(norm=norm, cmap=cmap),
- cax=ax, orientation='vertical', label='Some Units')
color in pymol
- from pymol import cmd
- num_classes = 100
- cmap = cm.rainbow(np.linspace(0.0, 1.0, num_classes))
- for i in range(num_classes):
- colorname='color%d'%(i)
- cmd.set_color(colorname, '[%f,%f,%f]'%(cmap[i][0],cmap[i][1],cmap[i][2]))
- cmd.delete("all")
- cmd.load(file)
- for index in df_normalized.index.values:
- resid = index + 1
- rmsf = df_normalized.iloc[index]
- colorid="color%d"%(int(float(rmsf)))
- cmd.color(colorid,"resi " + str(resid))
- cmd.save("test.pse")
