3.20 阳性化合物MD
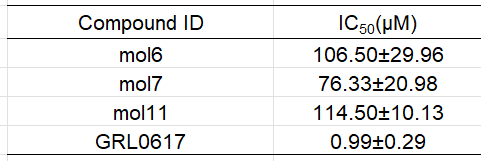
PL的阳性对照为GRL0617,复合物晶体结构 PDB:7JRN (分辨率最好)
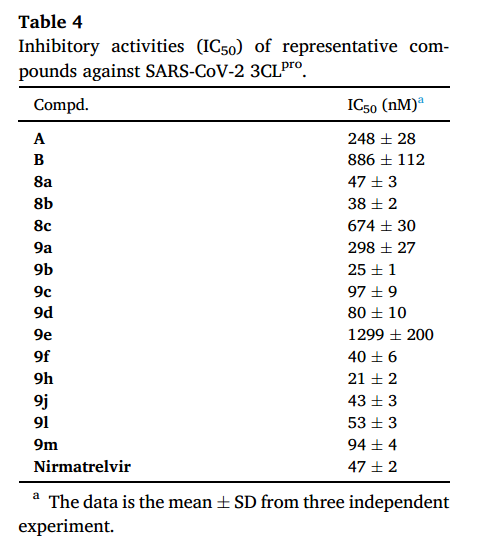
3CL的阳性对照为Nirmatrelvir,复合物晶体结构 PDB:7VH8
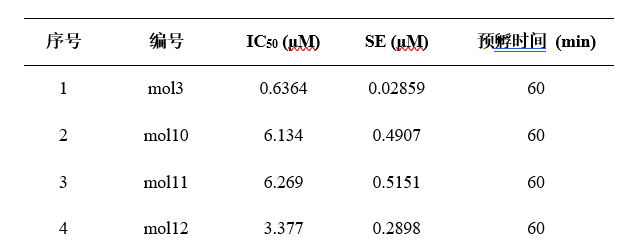
蛋白准备(晶体结构)
- 85.23 /home/databank_70t/zzy/project/hsbd/active_control/sys-pre/pro/pdb2pqr
- pdb2pqr30 ../7cmd.pdb 7cmd.pqr --ff AMBER --ffout AMBER --with-ph 7.4 --pdb-output 7cmd_complex.pdb
- grep -v HETATM 7cmd_complex.pdb |awk '$5=="A"{print}' >7cmd_apo_clean.pdb
- pdb2pqr30 ../7vh8.pdb 7vh8.pqr --ff AMBER --ffout AMBER --with-ph 7.4 --pdb-output 7vh8_complex.pdb
- grep -v HETATM 7vh8_complex.pdb >7vh8_apo_clean.pdb
配体准备
坐标信息(PDB)
质子化预测
- 23 /home/databank_70t/zzy/project/hsbd/active_control/sys-pre/ligand/ligprep
- for a in
`cat index`;do ~/software/Schrodinger_Suites_2021-2/ligprep -R e -epik -ph 7.4 -pht 0.0 -isd ../${a}.sdf -osd ./${a}.sdf;done - mv 7vh8_B_4WI.sdf nirmatrelvir.sdf
- mv 7cmd_E_TTT.sdf GRL0617.sdf
高斯优化
- 23 /home/databank_70t/zzy/project/hsbd/2025-4bioactive/sys-pre/ligand/g16
- # 使用antechamber生成配体小分子参数(85.23)
- for i in
`cat index`;do mkdir $i;cd $i;antechamber -i ../../ligprep/${i}.sdf -fi sdf -o $i.gjf -fo gcrt;sed -i '1,3d' $i.gjf;sed -i '1i %mem=4GB\n%nproc=4\n#B3LYP/6-31G* Pop=MK iop(6/33=2) iop(6/42=6) opt' $i.gjf;cd ../;done - # 提交 高斯优化任务 10:33
- 23 /home/databank_70t/zzy/project/hsbd/2025-4bioactive/sys-pre/ligand/g16
- for i in
`cat index`;do cd $i;antechamber -i $i.log -fi gout -o $i.prep -fo prepi -c resp;parmchk2 -i $i.prep -f prepi -o $i.frcmod -a y;cd ../;done
复合物体系准备
- 23 /home/databank_70t/zzy/project/hsbd/active_control/sys-pre/ligand/atom_name_check
- cp ../g16/index ./
- sh do.sh
- # 可视化检查
tleap
- # 23 /home/databank_70t/zzy/project/hsbd/active_control/sys-pre/tleap
- # 加载环境
- source ~/zzy/ff19sb.sh
- for i in
`cat index`;do mkdir -p $i;done - cp ../ligand/g16/GRL0617/*frcmod ./pl-monomer-7cmd/lig.frcmod
- cp ../ligand/g16/GRL0617/*prep ./pl-monomer-7cmd/lig.prep
- cp ../ligand/atom_name_check/GRL0617/LIG.PDB ./pl-monomer-7cmd/lig.pdb
- cp ../pro/pdb2pqr/7cmd_apo_clean.pdb ./pl-monomer-7cmd/pro.pdb
- cp ../ligand/g16/nirmatrelvir/*frcmod ./3cl-monomer-7vh8/lig.frcmod
- cp ../ligand/g16/nirmatrelvir/*prep ./3cl-monomer-7vh8/lig.prep
- cp ../ligand/atom_name_check/nirmatrelvir/LIG.PDB ./3cl-monomer-7vh8/lig.pdb
- cp ../pro/pdb2pqr/7vh8_apo_clean.pdb ./3cl-monomer-7vh8/pro.pdb
- for i in
`cat index`;do cp md_parm_gen_chinese.py $i;cd $i;python md_parm_gen_chinese.py 1;cd ../;done - # 可视化检查
amber_to_gmx
- # 23 /home/databank_70t/zzy/project/hsbd/active_control/sys-pre/amber_to_gmx
- #################################### do.sh #############################################
- for i in
`cat index` - do
- mkdir -p $i
- cp ../tleap/$i/complex* $i
- cd $i
- cp ../amber_to_gmx-add_restraint.py ./
- python amber_to_gmx-add_restraint.py 0 complex.prmtop complex.inpcrd
- python amber_to_gmx-add_restraint.py 1
- (echo '1|13'; echo 'q') | gmx_mpi make_ndx -f gmx.gro -o index.ndx
- echo 1|gmx_mpi genrestr -f gmx.gro -n index.ndx -o posre1.itp
- cd ../
- done
- ############################################################################################
- # 根据配体原子数生成 posre2.itp (原子数+4=行数)
- sed -n '1,47p' pl-monomer-7cmd/posre1.itp > pl-monomer-7cmd/posre2.itp
- sed -n '1,73p' 3cl-monomer-7vh8/posre1.itp > 3cl-monomer-7vh8/posre2.itp
预平衡
- # 8-4090(75.3) /home/dddc/zyzhou/project/hsbd
- scp -r dddc@172.21.85.23:/home/databank_70t/zzy/project/hsbd/active_control ./
- # min-nvt-npt.sh 在mdp文件合适的情况下仅需要修改工作路径,就可以自动完成所有体系的预平衡
- dddc@gpu-4090:/home/data/zzy/project/hsbd/2025-4bioactive/pre-equ$ nohup bash min-nvt-npt.sh &
- [1] 2697051
MD模拟任务
3cl-monomer-7vh8
- 75.3 /home/dddc/zyzhou/project/hsbd/active_control/cmd/100ns/1486-70-0-dock-PL
pl-monomer-7cmd
- 75.3 /home/dddc/zyzhou/project/hsbd/active_control/cmd/100ns/56083-03-5-dock-3CL
任务均已提交 2025-03-20