4.9 md-write
Initial structures for molecular dynamics simulation
比较活性位点对接结果和D3DOCK对接结果
- 75.2 /home/databank/zyzhou/project/hsbd/known-pkt
- (base) dddc@SR665-V3:/home/databank/zyzhou/project/hsbd/known-pkt$ grep -A 1 min */sys-pre/ligand/dock/*
- 3cl/sys-pre/ligand/dock/smina_56083-03-5-dock-3CL.sdf:> <minimizedAffinity>
- 3cl/sys-pre/ligand/dock/smina_56083-03-5-dock-3CL.sdf--7.67074
- --
- 3cl/sys-pre/ligand/dock/smina_59870-65-4-dock-3CL.sdf:> <minimizedAffinity>
- 3cl/sys-pre/ligand/dock/smina_59870-65-4-dock-3CL.sdf--7.91815
- --
- pl/sys-pre/ligand/dock/smina_1486-70-0-dock-PL.sdf:> <minimizedAffinity>
- pl/sys-pre/ligand/dock/smina_1486-70-0-dock-PL.sdf--7.90081
- --
- pl/sys-pre/ligand/dock/smina_56083-03-5-dock-PL.sdf:> <minimizedAffinity>
- pl/sys-pre/ligand/dock/smina_56083-03-5-dock-PL.sdf--9.61422
- --
- pl/sys-pre/ligand/dock/smina_91433-17-9-dock-PL.sdf:> <minimizedAffinity>
- pl/sys-pre/ligand/dock/smina_91433-17-9-dock-PL.sdf--7.90676
绘制初始结构比较图,展示对接打分
D3DOCK的对接中,要仔细想想 二聚体/三聚体的氨基酸序号
先全部按照Tleap-MD的编号来,后面看情况把第二条链上的序号减去第一条链(经检查,是完全一致的)
PL_dimer 314*2 314ILE
Dynamic behavior of protein-ligand complex
rmsd,rmsf都在这儿展示
75.3 的数据基本都转移到75.2了
- 75.2 /home/databank/zyzhou/project/hsbd/4bioactive-d3dock/analysis/rmsd/complex
- # 之前的图片都是CAS号命名的,要转成TCMID
图已有,要看怎么描述
google scholar"rmsd source:Journal source:of source:the source:American source:Chemical source:Society"
看看高水平杂志怎么描述的
rmsf同理
mmgbsa,rmsd.rmsf已整理好
mmgbsa t检验的单双尾,type要在verify一下
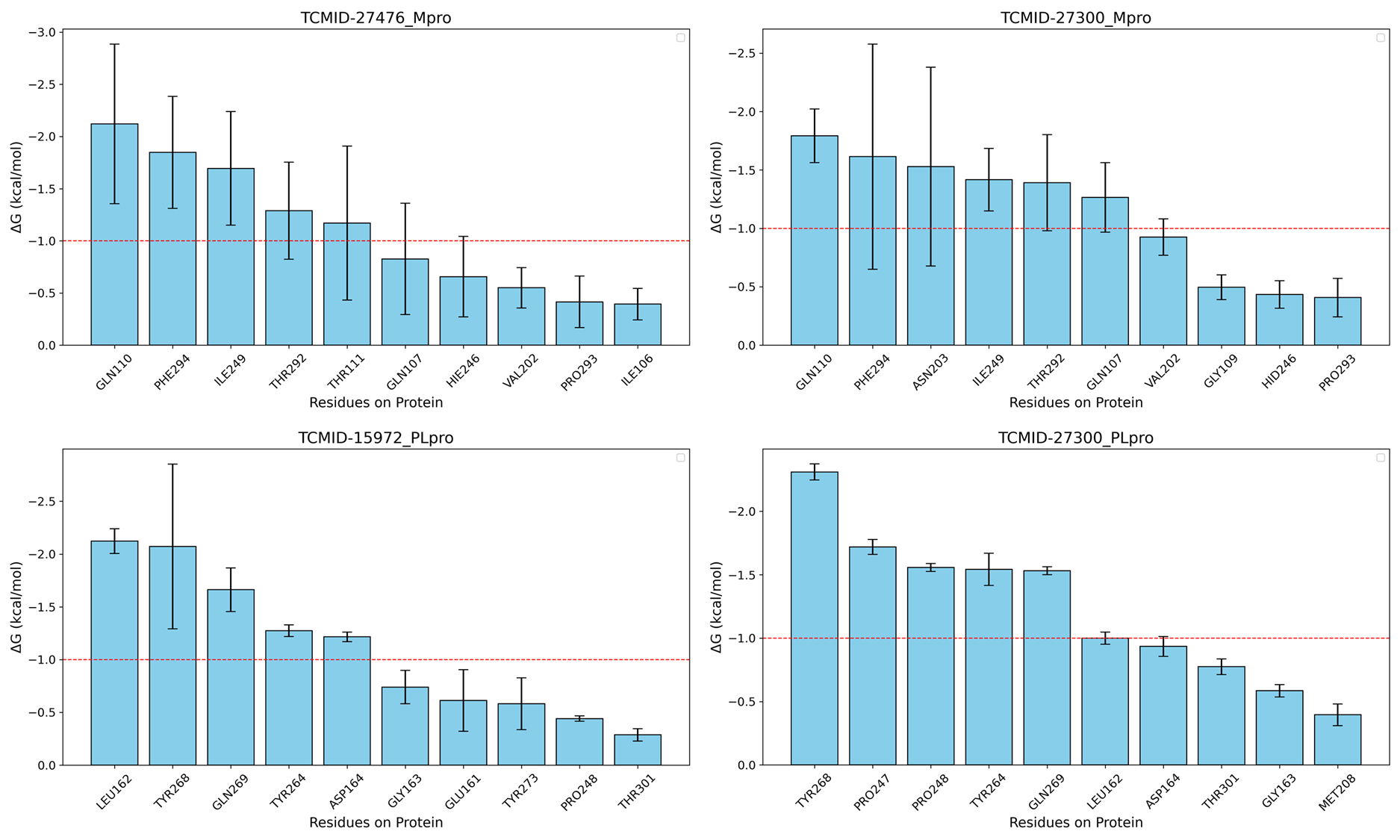
还差残基能量分解和残基接触频率
- 23 /home/databank_70t/zzy/project/hsbd/D3Targets-dock/workdir/5bioactive-dock-all-3cl_pl/analysis
就单纯的给出5个体系MD模拟较稳定的这个结论
Predicted highly frequent residues of protein-ligand binding
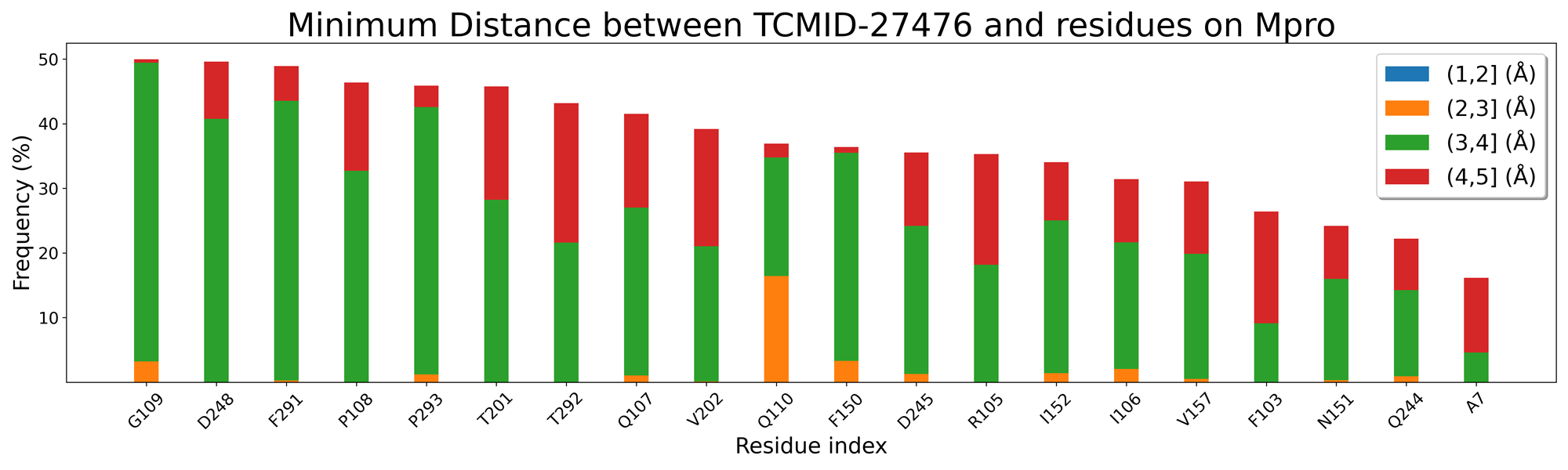
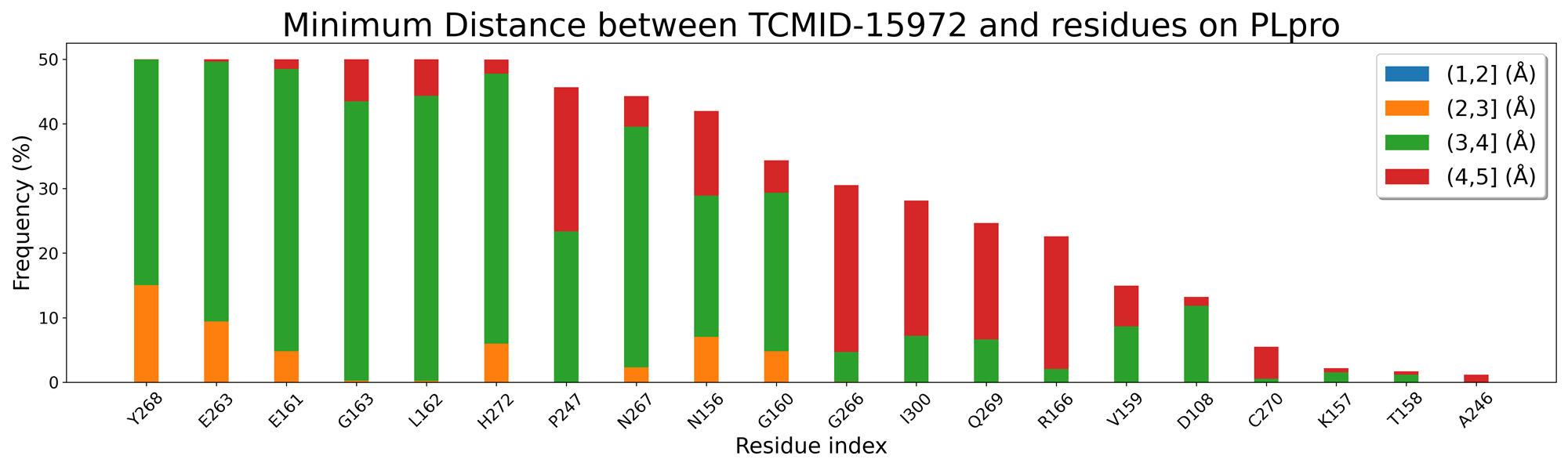
TCMID-27476 and TCMID-27300 MD中结合位点的证据:
AT7519-Mpro 复合物 ( PDB ID : 7AGA) X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease | Science
ARG298 这个残基有点意思,可展开讨论
针对AT7519-Mpro 复合物做的MD模拟,可参考:
Mpro 可药位点的一些讨论:
PLpro也是类似的思路:
找该活性位点的关键氨基酸,是否在高接触频率的残基中,以此展开讨论。
- 75.2 /home/databank/zyzhou/project/hsbd/4bioactive-d3dock/analysis/cluster/ligand_rmsd
- # 做了根据配体RMSD的聚类