0922 SMD 参数测试
- /home/databank/zzy/project/MD/koff/smd/cv-res_bb-mol-restbb-0918/trypsin-benzamidine/0922
测试一下 pull_coord1_rate ;pull_coord1_k 两个参数
根据文献中的图,
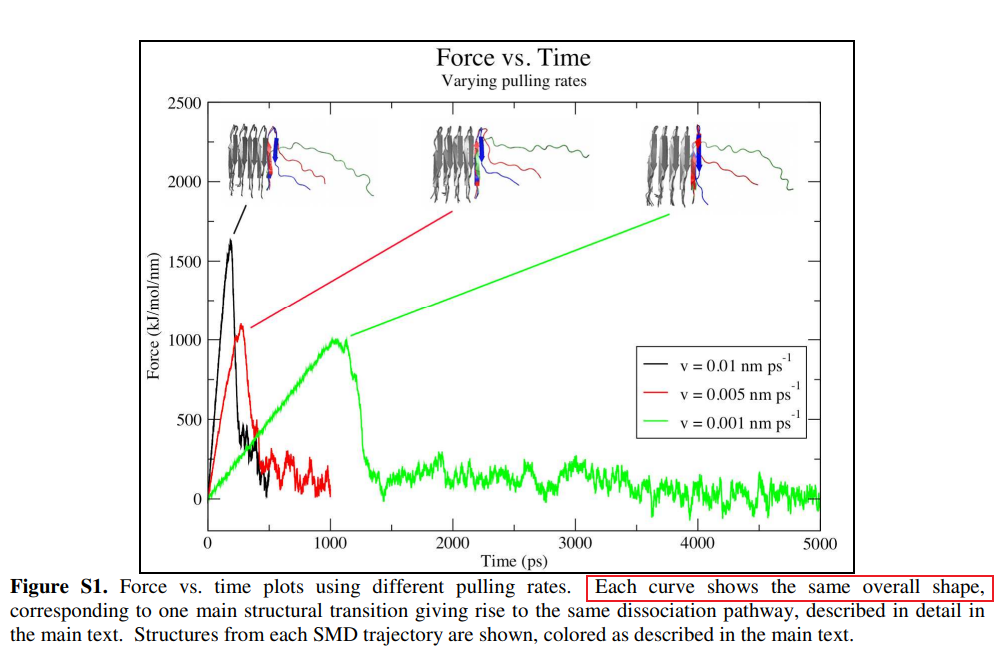
做一个类似的图出来
- /home/databank/zzy/project/MD/koff/smd/cv-res_bb-mol-restbb-0918/trypsin-benzamidine/0922
- # tpr
- gmx_mpi grompp -f smd.mdp -p ../../../params/gmx.top -c ../../../params/npt.gro -r ../../../params/npt.gro -n ../../../params/index.ndx -o smd.tpr -maxwarn 1
- nohup mpirun -np 50 gmx_mpi mdrun -v -deffnm smd &
- # 4090
- nohup mpirun gmx_mpi mdrun -v -deffnm smd &
- # vis 检查轨迹
- gmx_mpi trjconv -f ../smd.xtc -s ../smd.tpr -n ../../params/index.ndx -o dry_traj.pdb -pbc mol -ur compact
- sort -g -k2nr ../smd_pullf.xvg |head
- # cpptraj rms 一下,跟 dry.pdb 比较关键氨基酸位置差异
- # 85.24
- # /home/databank/zzy/project/MD/koff/smd/cv-res_bb-mol-restbb-0918/trypsin-benzamidine/0922/test-rate
- for i in
`cat index`;do gmx_mpi trjconv -f $i/smd.xtc -s $i/smd.tpr -n ../../params/index.ndx -o $i/dry.xtc -pbc mol -ur compact;done - for i in
`cat index`;do gmx_mpi trjconv -f $i/smd.xtc -s $i/smd.tpr -n ../../params/index.ndx -o $i/dry.pdb -pbc mol -ur compact -dump 0;done - # 再用gmx计算下dis
- for i in
`cat index`;do gmx_mpi distance -f ../$i/dry.xtc -s ../$i/dry.pdb -n ../../../params/index.ndx -select 'com of atomnr 2465 2467 2475 plus com of resid 225' -oall $i-dis-keyresnbb-MOL.xvg;done - paste *xvg>dis-keyresnbb-MOL.xvg
